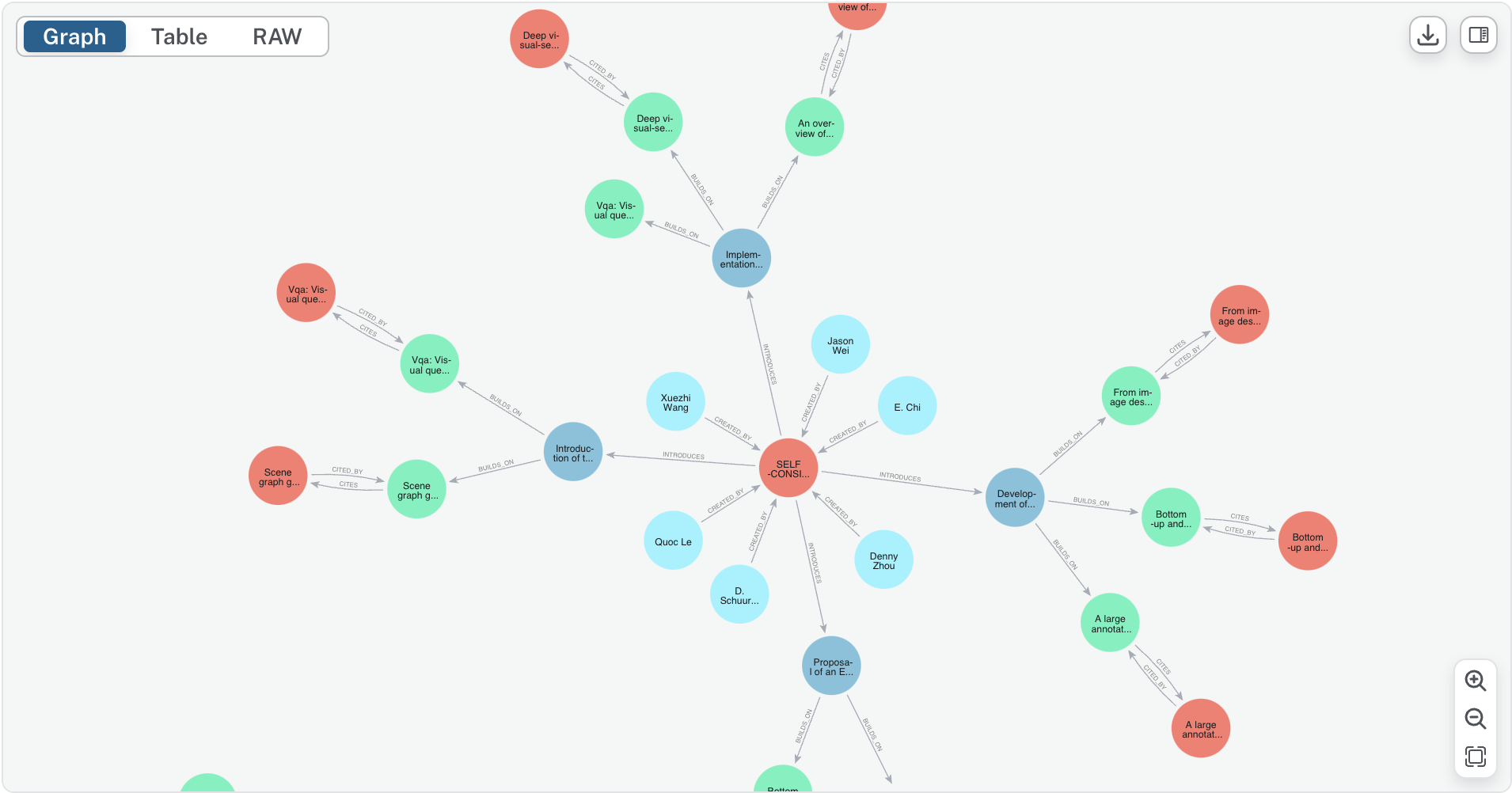
Innovation lineage tracking is the process of mapping out how ideas and inventions evolve over time, tracing each innovation’s roots back to the foundational works that influenced it. Much like tracing a family tree, innovation lineage tracking reveals the ancestry of ideas, showing how they build upon or diverge from one another over time. This method goes beyond traditional citation analysis by capturing the specific advancements each work introduces, providing a nuanced understanding of the innovation landscape.
AI-Enhanced Lineage Discovery combines automation with deep contextual analysis, using AI to extract, analyze, and link innovation details across research works. This approach highlights how AI not only automates the extraction of innovation insights but also enhances the understanding of complex interdependencies across research fields. By utilizing models like ColPali and Qwen in conjunction with Vespa for a “chat-with-document” setup, the system offers an interactive exploration of research papers. This includes generating Cypher queries with language models, providing structured, queryable insights through Neo4j and expanding to sources like Semantic Scholar and internet search. This multi-modal, multi-model setup creates a robust framework for innovation discovery, enabling more adaptive, wide-ranging lineage traversal through interconnected sources.
Use Cases and Applications of Innovation Lineage Tracking
Innovation lineage tracking serves various purposes, such as identifying foundational work, assessing novel contributions, and providing orientation for newcomers to a field. While this approach focuses on scientific research papers, it has significant applications in other areas like patent analysis, intellectual property, and business strategy, providing a structured roadmap for understanding how ideas have developed, diverged, and impacted their domains over time.
Here’s how AI-powered lineage tracking drives insight across different dimensions:
-
AI-Augmented Innovation Insights: AI models analyze text and image data to identify, categorize, and contextualize key innovations, providing structured insights that reveal each contribution’s significance and purpose within its field.
-
Lineage Mapping and Impact Tracing: By using AI to detect explicit citations and lineage, the system clarifies how each innovation builds on prior work or diverges from it. This lineage mapping provides a comprehensive view of innovation pathways, making it easier to trace a concept’s evolution and assess its impact over time.
-
Intelligent Contextual Analysis: AI leverages contextual understanding to distinguish true innovation from incremental advancements. By classifying relationships between innovations and past work, it helps prioritize influential sources and illuminate critical junctures in knowledge development.
-
Scalable Knowledge Synthesis: AI scales up the traditionally manual process of lineage tracking, producing deep insights at a fraction of the time, even across expansive bodies of research. This scalability enables a holistic view of trends, clusters of influence, and emerging innovation hot spots.
This system’s multi-tiered, adaptive framework not only addresses traditional research but also extends to more diverse data sources. With each new innovation analyzed, AI-driven lineage tracking deepens the understanding of knowledge evolution, offering a reliable, intelligent assistant that continuously enriches the landscape of research insights.
Let’s look at some code for a view of how some of this might be implemented. We’ll call the tool InfluTrace.
Here’s a detailed description with code-level discussion based on your outline:
Overview of InfluTrace’s Functionality
InfluTrace is a specialized AI-powered research assistant designed to automate the extraction, analysis, and mapping of academic research innovations. By converting academic PDFs into structured lineage data, InfluTrace links each innovation to relevant prior works, constructing a detailed lineage that shows how ideas evolve and interconnect. Here’s a breakdown of its main functions:
- PDF Processing and Text Extraction
- Code Overview: InfluTrace employs
pdf2imageandpypdflibraries to extract text and images from PDFs, segmenting content for detailed analysis. Theget_pdf_images()function, for example, processes each page by converting it to images for OCR (optical character recognition) tasks. Here’s a snippet:def get_pdf_images(pdf_url): pdf_file = download_pdf(pdf_url) images = convert_from_path("temp.pdf") # Converts PDF to images reader = PdfReader("temp.pdf") texts = [page.extract_text() for page in reader.pages] return images, texts - This step prepares each section of the paper for further processing by machine learning models, enabling precise detection of key sections like Abstract, References, and Methodology.
- Code Overview: InfluTrace employs
- Innovation Detection and Classification
- InfluTrace uses models like Google’s Gemini Flash Vision and OpenAI’s GPT to identify and classify core innovations within each paper. In the
extract_innovations()function, a series of LLM prompts are sent to classify each innovation as a concept, technique, application, or finding. - Here’s a simplified version of the core prompt logic:
def extract_innovations(query, images): response = gemini_model.generate_content([query, *images]) data, _ = extract_json_from_response(response.text) return data # Returns structured JSON containing detected innovations - By running these classification models over segmented content, the system ensures that innovations are correctly categorized, capturing each contribution’s significance and novelty.
- InfluTrace uses models like Google’s Gemini Flash Vision and OpenAI’s GPT to identify and classify core innovations within each paper. In the
- Lineage Analysis and Relationship Mapping
- InfluTrace then performs lineage analysis by matching references within each paper to its innovations. Each detected innovation is annotated with its influences, allowing the system to establish relationships like
BUILDS_ONandINTRODUCES. - The
process_pdf_doc()function consolidates these relationships in Neo4j:def process_pdf_doc(query, images, texts, url): innovations = extract_innovations(query, images) for innovation in innovations["innovations"]: lineage_data = extract_related_references(innovation, texts) store_paper_with_relationships(lineage_data) # Stores in Neo4j - This function stores key data like innovation descriptions and lineage sources in a structured format, creating a rich network of interconnected research data.
- InfluTrace then performs lineage analysis by matching references within each paper to its innovations. Each detected innovation is annotated with its influences, allowing the system to establish relationships like
- Graph Database Modeling
- Neo4j is employed to store entities and relationships such as Papers, Authors, Innovations, and Lineages. For example, the
store_paper_with_relationships()function leverages Cypher queries to store relationships likeCITES,CITED_BY,INTRODUCES, andBUILDS_ON:def store_paper_with_relationships(paper_data): query = ("MERGE (p:Paper {paper_uuid: $paper_uuid}) " "SET p.title = $title " "MERGE (i:Innovation {innovation_uuid: $innovation_uuid}) " "MERGE (p)-[:INTRODUCES]->(i)") session.run(query, **params) # Executes in Neo4j - This graph structure allows for rapid querying of connected research work, enabling users to efficiently trace academic influences and track the evolution of innovations over time.
- Neo4j is employed to store entities and relationships such as Papers, Authors, Innovations, and Lineages. For example, the
- API Integration with Semantic Scholar
- InfluTrace retrieves additional citation metadata through the Semantic Scholar API, including author names and open-access PDF links. This ensures accurate citation matching and fills in missing data, allowing InfluTrace to maintain comprehensive citation records. Here’s a sample integration call:
def fetch_pdf_urls(citation, reference_info): response = requests.get("https://api.semanticscholar.org/graph/v1/paper/search", params=params) if response.status_code == 200: return response.json().get("data", [])[0]
- InfluTrace retrieves additional citation metadata through the Semantic Scholar API, including author names and open-access PDF links. This ensures accurate citation matching and fills in missing data, allowing InfluTrace to maintain comprehensive citation records. Here’s a sample integration call:
- Error Handling and Data Validation
- InfluTrace includes a robust error-handling framework that ensures reliable results. By applying an Adaptive Multi-Tiered Retry Framework, InfluTrace retries LLM requests in multiple ways (e.g., simple retries, retries with prompt rewriting, model substitution) when errors occur. Additionally, every response undergoes data validation to ensure it conforms to the expected JSON structure.
Technical Highlights
-
Machine Learning Models: InfluTrace leverages state-of-the-art models like ColPali for embedding document content and OpenAI models for natural language processing, ensuring high accuracy in text and citation extraction tasks.
-
Graph Relationships and Semantic Querying: Neo4j’s graph structure provides a flexible querying layer for connected data, making it easy to trace academic influences and innovation evolution. The use of structured relationships like
CITESandBUILDS_ONenables powerful, semantically rich exploration of data. -
Data Reliability: InfluTrace employs advanced LLM prompts with context-driven retries to ensure that only directly mentioned citations are included in lineage data, reducing the risk of irrelevant or implied references.
Data Structures for Organizing Innovation Insights and Lineage
At the core of InfluTrace’s functionality is a structured, JSON-based representation of each research paper’s key innovations, relationships, and influence paths. This structured data format allows for precise lineage mapping and seamless integration into a Neo4j knowledge graph, where relationships among innovations can be queried and analyzed effectively. Below is an overview of the main data structure components:
-
Innovation Data Structure
Each innovation within a paper is represented as a structured dictionary, with key attributes including
innovation_id,description,type, andpurpose. This provides a uniform schema for capturing the essence of each innovation. For example:{ "innovation_id": "1", "description": "Introduction of a novel MTL architecture termed “Slice Networks” for deep neural networks.", "type": "New Concept", "purpose": "Offers a generalized approach to MTL in deep learning, allowing for flexible sharing of network components across tasks, potentially enhancing efficiency and improving generalization.", } -
Lineage Tracking and Relationship Mapping
Each innovation has a
lineagefield that captures its connections to prior works. Each lineage item includes:citation: The precise citation as referenced in the paper.reference: The full bibliographic information of the cited work.contributions: A list summarizing the cited work’s relevant contributions.relationship: Describes how the innovation builds on, diverges from, or reinterprets the cited work.url: Links to the full-text source, enhancing traceability and access.
For instance, a lineage item might look like this:
{ "citation": "Baxter_1997", "reference": "Baxter, J. (1997). A Bayesian/information theoretic model of learning to learn via multiple task sampling. Machine Learning, 28, 7–39.", "title": "A Bayesian/information theoretic model of learning to learn via multiple task sampling", "contributions": ["early work on MTL in neural networks, focusing on hard parameter sharing"], "relationship": "This innovation extends Baxter's work by introducing a more flexible structure for sharing parameters, enabling finer control over the shared and task-specific layers.", "url": "https://link.springer.com/content/pdf/10.1023/A:1007327622663.pdf", "semantic_scholar_id": "1bd6e929ed8384ea2212d50ab3c103ec018cc9fd" } -
Graph Database Modeling
Once extracted, these innovations and lineage relationships are stored in Neo4j, with distinct nodes for Papers, Innovations, Authors, and Lineages. Key relationships include:
- CITES and CITED_BY: Connecting papers that reference one another.
- INTRODUCES: Linking each paper to its innovations.
- BUILDS_ON: Showing how each innovation is influenced by prior works.
-
Use Cases
This structured approach allows InfluTrace to:
- Enable Targeted Lineage Queries: Users can trace the development of specific innovations by querying
BUILDS_ONrelationships. - Support Comprehensive Analysis: The inclusion of details like
contributionsandrelationshipallows for nuanced lineage analysis, highlighting not just connections but the nature of each influence. - Enhance Accessibility: With
urlfields and Semantic Scholar IDs, users can easily access cited works, deepening their understanding of the innovation’s foundation.
- Enable Targeted Lineage Queries: Users can trace the development of specific innovations by querying
We’ll look next at document chat, so stay tuned.